Assignment of 1H-NMR spectra
On this page we will deal with how to interpret an NMR spectrum. The meaning of assignment in the title is to assign each peak to a proton in the molecule under investigation. The examples here are of 1D proton assignments. For more complex examples, see the 2D assignments of 12,14-ditbutylbenzo[g]chrysene and cholesteryl acetate.
In the example in fig. 1 of isopropyl-β-D-thiogalactopyranoside (shown without the hydrogens for simplicity – each carbon has four bonds, click here to see the molecule with hydrogens), all the hydroxyls have been exchanged with the deuterium oxide solvent to deuteroxyls. Therefore, the hydroxyl signals do not appear in the spectrum and do not couple with the other signals, making the spectrum simpler.
Fig. 1. 1H-NMR spectrum of isopropyl-β-D-thiogalactopyranoside in D2O
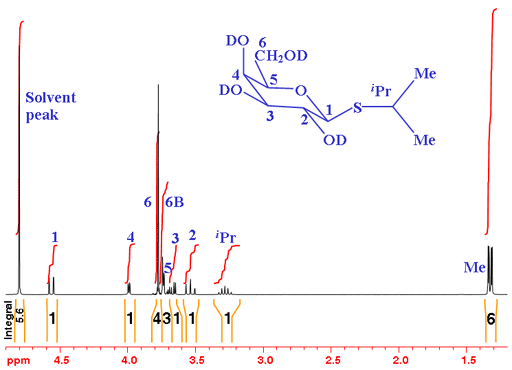
From the integrals, we see that there are two multiples of three, one of which has tall sharp signals so very likely corresponds to the two methyl (CH3) signals. The remaining signals are expected to yield integrals of one so the integrals of three and four are overlapping. H6 is expected to yield two separate signals because they are diasteriomeric (if one of them is exchanged with another group, the attached carbon would be optically active. This fact affects their chemical shift and they differ magnetically - If you don't understand this, don't worry, just take it form granted for now).
From the chemical shifts we see that what we suspect are methyls have the appropriate chemical shift and the remaining signals fall in the overlapping CH and CH2 regions as expected. If you are an experienced sugar chemist you will know that the signal with the highest chemical shift is usually the anomeric signal (H1 – the hydrogen connected to the carbon next to the sugar ring-closing oxygen).
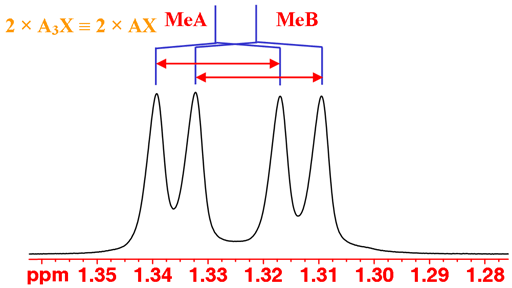
The coupling patterns can be used to continue the analysis. You could be forgiven for thinking that the methyl signals display an AXY coupling pattern. However, they only couple with the single iPr proton so should yield an AX pattern. The reason is that the methyls (labeled MeA and MeB) are diasteriomeric so have different chemical shifts (not magnetically identical, just like the H6 protons). The result is two overlapping AX patterns (fig. 2).
Fig. 2. The methyl doublets of isopropyl-β-D-thiogalactopyranoside in D2O
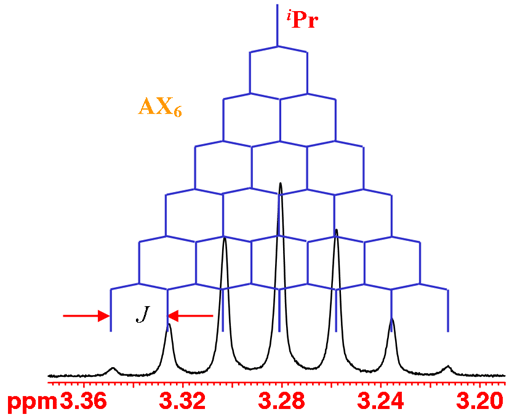
The iPr proton is coupled to six methyl protons yielding a septet (fig. 3).
Fig. 3. The iPr septet of isopropyl-β-D-thiogalactopyranoside in D2O
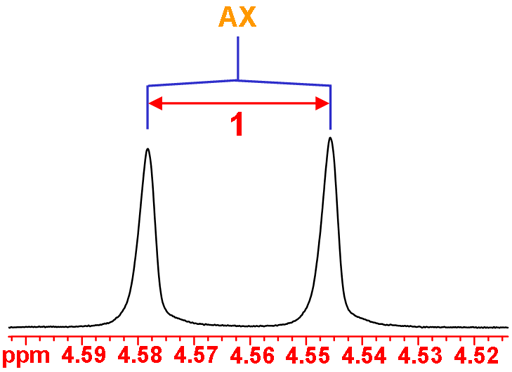
The anomeric H1 is coupled to H2 yielding an AX doublet (fig. 4).
Fig. 4. The anomeric H1 doublet of isopropyl-β-D-thiogalactopyranoside in D2O
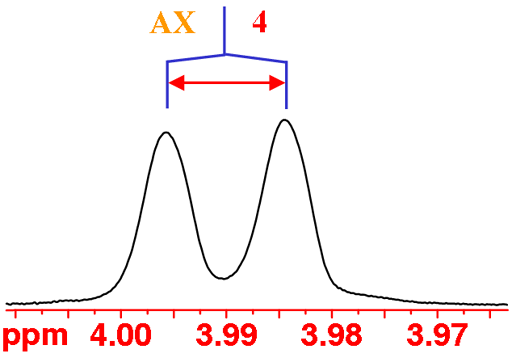
H4 has an unusually small coupling to H5 (this occurs when the two CH bonds are approximately at right-angles to each other), so small that it is not observed in a normal spectrum. So H4 displays an AX pattern instead of the expected AXY pattern although the peaks are slightly broad indicating the missing coupling (fig. 5).
Fig. 5. The H4 multiplet of isopropyl-β-D-thiogalactopyranoside in D2O
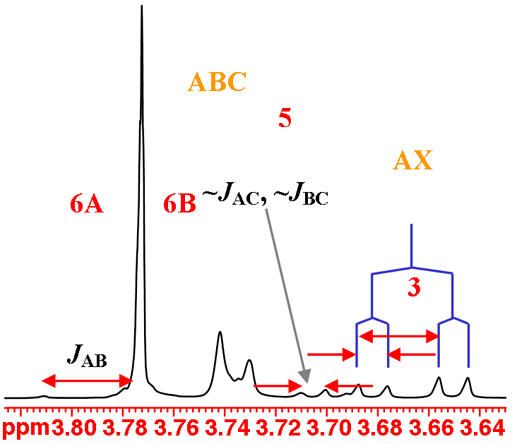
H3 couples with both H2 and H4 and yields the expected AXY pattern. While H5, H6A and H6B have very similar chemical shifts and stong coupling that combine to yield very strongly second order coupled ABC pattern that is difficult to analyze (fig. 6).
Fig. 6. The H3 and the overlapping H5, H6A and H6B multiplets of isopropyl-β-D-thiogalactopyranoside in D2O
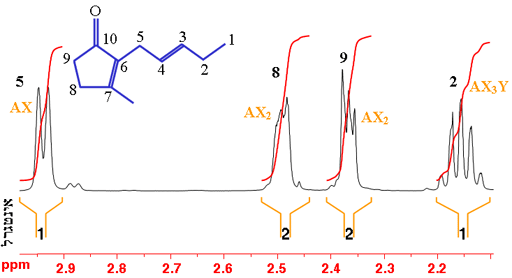
In the example of trans-geraniol in fig. 7 (shown without the hydrogens for simplicity – each carbon has four bonds, click here to see the molecule with hydrogens), proton-5 (H5) is coupled and therefore split by proton-4 (H4); H8 and H9 represent two protons each that are split by each other into triplet AX2 patterns; and H2 is split into four by the three protons at H1 and the resulting quartet is split again by H3. However, second order coupling distorts the multiplets making the assignment more difficult.
Fig. 7. Part of the 1H-NMR spectrum of trans-geraniol in CDCl3